Researchers Discover Two, Rare Genes Associated with Alzheimer’s Disease
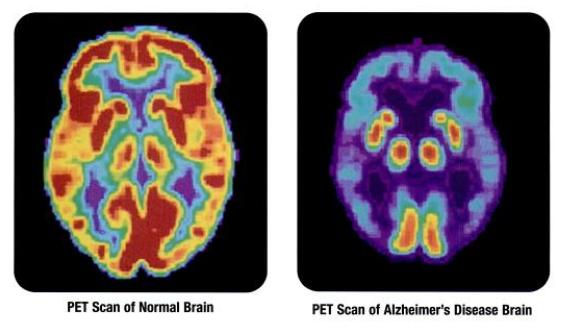
PET scans showing the differances between a normal older adult's brain and the brain of an older adult afflicted with Alzheimer's disease. Source: public domain
Source: Boston University School of Medicine Redistributed by NHI, April 1, 2019
Researchers have identified two, extremely rare genetic variants linked to Alzheimer disease (AD) for the first time.
These variants, one located in the NOTCH3 gene and the other in the TREM2 gene, were observed in persons with AD but not in any of the controls.
According to the researchers, the NOTCH3 variant has not been implicated in AD in previous large genetic studies, however, other mutations in this gene cause a very rare form of dementia called CADASIL which begins with severe headaches and strokes in young adulthood followed by dementia by midlife (decades before the typical age when late-onset AD occurs).
Other mutations in the TREM2 gene have been associated with AD, and it was previously shown that persons who carry two copies of this particular mutation (referred to as Q33X) have a very rare disorder called Nasu-Hakola disease which is characterized by onset of dementia in midlife and polycystic bone lesions with fractures.
Although the NOTCH3 mutation causing AD is very rare in virtually all racial and ethnic groups, it is much more frequent in Ashkenazi Jews, and the researchers determined that nearly all of the AD cases with the NOTCH3 mutation were of that descent.
“Our findings indicate that different mutations in the same gene or different number of copies of a particular mutation may lead to very distinct forms of dementia,” explained corresponding author Lindsay Farrer, PhD, chief of the Biomedical Genetics division. “Discovery of associations of Alzheimer’s risk with rare genetic variants can lead to new insights about biological pathways involved in AD and strategies for developing novel treatments and biomarkers.”
These findings emerged from analyses of the entire DNA sequence for the portions of the genome that encode genes (called exons) of more than 5,600 participants with AD and nearly 4,600 cognitively healthy elderly controls. DNA sequence data were generated by the Alzheimer Disease Sequencing Project, a large NIH-funded initiative that emerged from the National Alzheimer’s Act of 2012, and screened for variants that were present in persons with AD but not in the controls. The researchers also showed that study participants with AD compared to controls had a significantly greater burden of mutations in genes known to have a role in AD.
Dr. Farrer believes that if the findings about the NOTCH3 mutation are confirmed in a large independent sample of Ashkenazi Jews, a diagnostic and predictive test for AD could be developed for that specific population.
These findings appear online in JAMA Network Open.
Funding for this study was provided in part by NIA grants R01-AG048927, P30AG13846 and RF1-AG057519. The Alzheimer’s Disease Sequencing Project (ADSP) is comprised of two Alzheimer disease genetics consortia and three National Human Genome Research Institute (NHGRI) funded Large Scale Sequencing and Analysis Centers (LSAC). The two AD genetics consortia are the Alzheimer’s Disease Genetics Consortium (ADGC) funded by NIA (U01-AG032984), and the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) funded by NIA (R01 AG033193), the National Heart, Lung, and Blood Institute (NHLBI), other National Institute of Health (NIH) institutes and other foreign governmental and non-governmental organizations. The Discovery Phase analysis of sequence data is supported by NIA grants UF1-AG047133, U01-AG049505, U01-AG049506, U01-AG049507, and U01AG049508. The Discovery Extension Phase analysis is supported by NIA grants U01AG052411, U01-AG052410, and U01- AG052409. Data generation and harmonization in the Follow-up Phases is supported by NIA grant U54-AG052427.