Effective Neutralization of SARS-CoV-2 by Single-Domain Camelid Antibodies (Nanobodies) Offers hope in Covid-19 Treatment

Dr. Prangya P.Tripathy, PhD, Neucrad Health Desk, July 31, 2020
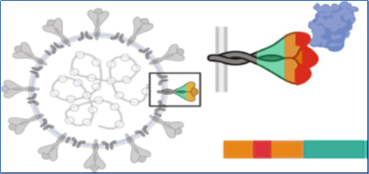
The curiosity for effective treatments to tackle COVID-19 pandemic lead researchers for the development of specific antibodies that can neutralize the SARS-CoV-2 virus responsible for COVID-19. Llama derived “nanobody” technology in this direction offers hope for use in the similar way to convalescent plasma therapy (Plasma therapy utilizes human antibodies present in the plasma of recovered Covid-19 person to treat the infected person). A recent study by Wrapp et al. (May, 2020) demonstrated binding affinity of “nanobodies” from Llama with Spike protein of SARS-COV-2 and effective neutralization of the virus. In a separate study (Huo et al., 2020), the researchers identified and characterized two closely related nanobodies H11-D4 and H11-H4, that could block the attachment of the SARS-CoV-2 spike to ACE2 (Angiotensin converting enzyme 2) in cell culture.
About Coronaviruses
These are enveloped positive-sense RNA viruses and are divided into four genera (α, β, g, and d). They are capable to infect a wide variety of host organisms including humans. Human disease causing corona viruses belong to seven categories, and four of these viruses (HCoV-HKU1, HCoV-OC43, HCoV-NL63, and HCoV-229E) circulate seasonally throughout the global population, causing mild respiratory disease in most patients. The three remaining viruses, SARSCoV-1, MERS-CoV, and SARS-CoV-2, are zoonotic pathogens that caused the epidemics or pandemics with severe to fatal symptoms in human population.
What are Nanobodies
Along with normal antibodies, camelids, such as llamas also produce heavy chain antibodies (HCAbs) which contain a single variable domain (VHH) as opposed to two variable domains (VH and VL). This single variable domain, in the absence of an effector domain, is referred to as a single-domain antibody, VHH or Nanobody and typically can have better affinities and specificities for antigens as compared to conventional antibodies. VHHs can be easily constructed into multivalent formats. Due to their thermal stability and chemostability, they are less susceptible to steric hindrances as compared to larger conventional antibodies. Valuable biophysical properties of several VHHs have led to the evaluation of nanobodies as therapeutics against common respiratory pathogens e.g. respiratory syncytial viruses (RSV). Because of the highly stable nature of VHHs, they attract special application in the context of respiratory infections where they can be nebulized and administered via an inhaler directly to the site of respiratory infections. Further, because of their stability they could be stored as therapeutic treatment options to tackle any epidemic.
Spike Proteins in CoronaVirus
Corona virus is covered with a spike(S) glycoprotein (a large class 1 fusion protein) and forms a trimeric complex. This complex can be functionally categorized into two distinct subunits, S1 and S2, which are separated by a protease cleavage site. The receptor-binding domain (RBD) is located in S1 subunit, which interacts with a host cell receptor protein to trigger membrane fusion. The S2 subunit contains the membrane fusion machinery, including the hydrophobic fusion peptide and the α-helical heptad repeats. The functional host cell receptors for SARS-CoV-1 is angiotensin converting enzyme 2 (ACE2) and for MERS-CoV is dipeptidyl peptidase 4 (DPP4), respectively. The SARS-CoV-2 S proteins also use ACE2 as a functional host-cell receptor, and several structures of this complex have already been reported. Neutralization mechanisms of anti-SARS-CoV-1 RBD and anti-MERS-CoV RBD antibodies is attributed to the blockage of the receptor-binding site and to trapping the RBD in the unstable up conformation. This leads to gradual destabilization of S1 and triggered to S2 to start fusion of the membrane prematurely.
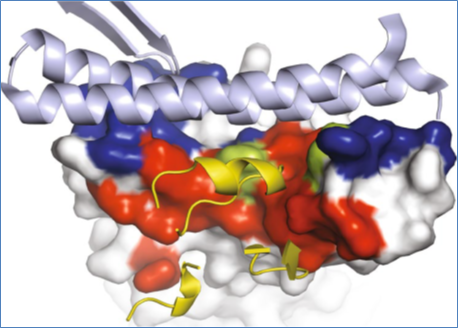
Interaction of Nanobodies with SARS-CoV2
Although many therapeutic antibodies are in use, nanobodies have recently entered into clinical trials, with a licensed one caplacizumab. A recent study, published in the Journal Nature Structural & Molecular Biology, reported identification and characterization of two closely related nanobodies H11-D4 and H11-H4, that could block the attachment of the SARS-CoV-2 spike to ACE2 in cell culture. Both the nanobodies recognize the same epitope, which partly overlaps with the ACE2 binding surface, explaining the blocking of the RBD–ACE2 interaction. The Nanobody-Fc fusions explained neutralizing activity against SARS-CoV-2 (4–6 nM for H11-H4, 18 nM for H11-D4) and also demonstrated a better neutralization with the SARS-CoV-1/2 antibody CR3022 (Huo et al. 2020).
In a separate study Wrapp et al., 2020 reported the isolation of two potently neutralizing VHHs directed against the SARS-CoV-1 and MERS-CoV RBDs, respectively. These VHHs were derived as an immunization response of the llama with prefused SARS-CoV-1 and MERS-CoV S proteins. The crystal structures of these two VHHs in complex with their respective viral epitopes were solved, which suggested likely mechanisms of neutralization. They also reported that the SARS-CoV-1 RBD directed VHH cross-reacts with the SARS-CoV-2 RBD and can block the receptor-binding interface. After engineering this VHH into a bivalent Fc-fusion, the cross-reactivity of VHH was checked and also neutralizes SARS-CoV-2 S pseudoviruses. Thus VHH-Fc fusion can be produced at high yields in an industry-standard CHO cell system that could serve as a potential therapeutic for the ongoing COVID-19 pandemic.
References
- Forsman, A., Beirnaert, E., Aasa-Chapman, M.M., Hoorelbeke, B., Hijazi, K., Koh, W., Tack, V., Szynol, A., Kelly, C., McKnight, A., et al. (2008). Llama antibody fragments with cross-subtype human immunodeficiency virus type 1 (HIV-1)- neutralizing properties and high affinity for HIV-1 gp120. J. Virol. 82, 12069–12081.
- Detalle, L., Stohr, T., Palomo, C., Piedra, P.A., Gilbert, B.E., Mas, V., Millar, A. Power, U.F., Stortelers, C., Allosery, K., et al. (2015). Generation and Characterizatio of ALX-0171, a Potent Novel Therapeutic Nanobody for the Treatment of Respiratory Syncytial Virus Infection. Antimicrob. Agents Chemother.
- Rossey, I., Gilman, M.S., Kabeche, S.C., Sedeyn, K., Wrapp, D., Kanekiyo, M.,Chen, M., Mas, V., Spitaels, J., Melero, J.A., et al. (2017). Potent single-domain antibodies that arrest respiratory syncytial virus fusion protein in its prefusion state. Nat. Commun. 8, 14158.
- Hoffmann, M., Kleine-Weber, H., Kru¨ ger, N., Mu¨ ller, M., Drosten, C., and Po hlmann, S. (2020). The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. bioRxiv. https://doi.org/10.1101/2020.01.31.929042.
- Peyvandi, F. et al. Caplacizumab reduces the frequency of major thromboembolic events, exacerbations and death in patients with acquired thrombotic thrombocytopenic purpura. J. Thromb. Haemost. 15 1448–1452 (2017)
- Wrapp, D. et al. Structural basis for potent neutralization of Betacoronaviruses by single-domain camelid antibodies. Cell 181, 1436–1441 (2020).
- De Vlieger, D., Ballegeer, M., Rossey, I., Schepens, B., and Saelens, X. (2018). Single-Domain Antibodies and Their Formatting to Combat Viral Infections. Antibodies (Basel) 8, 1.
- Detalle, L., Stohr, T., Palomo, C., Piedra, P.A., Gilbert, B.E., Mas, V., Millar, A., Power, U.F., Stortelers, C., Allosery, K., et al. (2015). Generation and Characterization of ALX-0171, a Potent Novel Therapeutic Nanobody for the Treatment of Respiratory Syncytial Virus Infection. Antimicrob. Agents Chemother. 60, 6–13.
- Wrapp, D., Wang, N., Corbett, K.S., Goldsmith, J.A., Hsieh, C.L., Abiona, O., Graham, B.S., and McLellan, J.S. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263.
- Bosch, B.J., van der Zee, R., de Haan, C.A., and Rottier, P.J. (2003). The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 77, 8801–8811.
- Hamers-Casterman, C., Atarhouch, T., Muyldermans, S., Robinson, G., Hamers, C., Songa, E.B., Bendahman, N., and Hamers, R. (1993). Naturally occurring antibodies devoid of light chains. Nature 363, 446–448.
- Huo, J., Bas A. ….Naismith J.H. (2020). Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nature Structural & Molecular Biology. . https://doi.org/10.1038/s41594-020-0469-6 ht-0tp








Nice informative article!
Thank you for going through it. Please explore http://www.neucradhealth.in